sylph - fast and precise species-level metagenomic profiling with ANIs
Sylph is a program that performs (1) metagenomic profiling or (2) containment average nucleotide identity querying for metagenomic shotgun sequencing samples.
-
Metagenomic profiling: sylph can determine the species/taxa in your sample and their abundances, just like Kraken or MetaPhlAn.
-
Containment ANI querying: sylph can search a genome, e.g. E. coli, against your sample. If sylph outputs an estimate of 97% ANI, your sample contains an E. coli with 97% ANI to the queried genome.
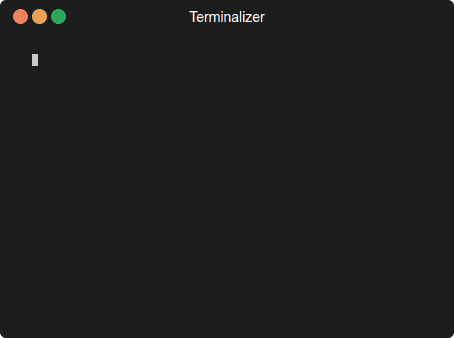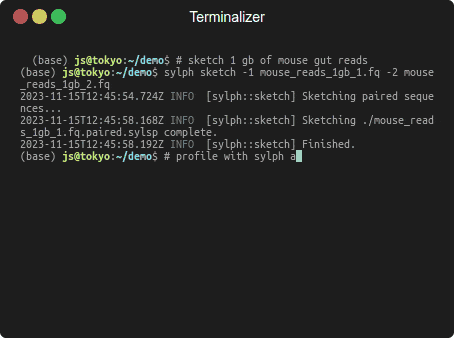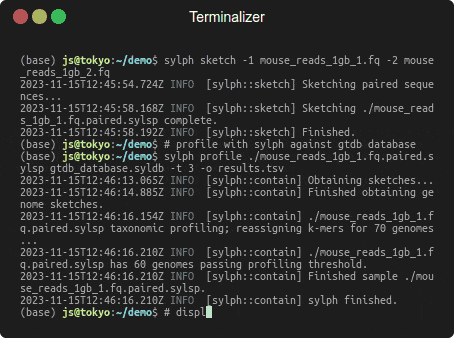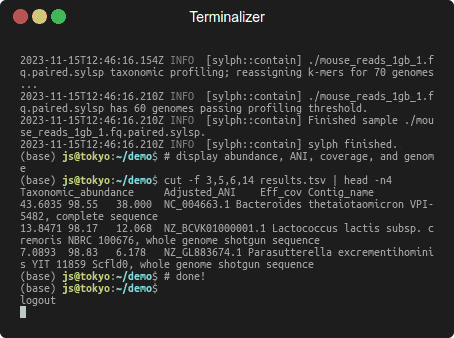
Profiling 1 Gbp of mouse gut reads against 85,205 genomes in a few seconds
Why sylph?
-
Precise species-level profiling: sylph has less false positives than Kraken and is about as precise and sensitive as marker gene methods (MetaPhlAn, mOTUs).
-
Ultrafast, multithreaded, multi-sample: sylph can be > 50x faster than other methods. Sylph only takes ~15GB of RAM for profiling against the entire GTDB-R220 database (110k genomes).
-
Accurate (containment) ANI information: sylph can give accurate ANI estimates between reference genomes and your metagenome sample down to 0.1x coverage.
-
Customizable databases and pre-built databases: We offer pre-built databases of prokaryotes, viruses, eukaryotes. Custom databases (e.g. using your own MAGs) are easy to build.
-
Short or long reads: Sylph was also the most accurate method on Oxford Nanopore's independent benchmarks.
How does sylph work?
sylph uses a k-mer containment method. sylph's novelty lies in using a statistical technique to estimate k-mer containment for low coverage genomes , giving accurate results for low abundance organisms. See here for more information on what sylph can and can not do.
How do I use sylph?
See the left side bar for more information. See the quick start and installation instructions for a quick start.
Changelog
See the CHANGELOG for complete details.
Citing sylph
Jim Shaw and Yun William Yu. Rapid species-level metagenome profiling and containment estimation with sylph (2024). Nature Biotechnology.